* Provision of complete systems
* Provision of bioinformatics software repositories
* Addition of bioinformatics packages to standard distributions
* Live DVD/CDs with bioinformatics software added
* Community building and support systems
There are now various projects with similar aims, on both Linux systems and other Unices, and a selection of these are given below. There is also an overview in the Canadian Bioinformatics Helpdesk Newsletter that details some of the Linux-based projects.
Package repositories.
Package repositories are generally specific to the distribution of Linux the bioinformatician is using. A number of Linux variants are prevalent in bioinformatics work. Fedora is a freely-distributed version of the commercial Red Hat system. Red Hat is widely used in the corporate world as they offer commercial support and training packages. Fedora Core is a community supported derivative of Red Hat and is popular amongst those who like Red Hat's system but don't require commercial support. Many users of bioinformatics applications have produced RPMs (Red Hat's package format) designed to work with Fedora, which you can potentially also install on Red Hat Enterprise Linux systems. Other distributions such as Mandriva and SUSE use RPMs, so these packages may also work on these distributions.
* Biolinux (Fedora)
* BioRPMs (RedHat and Fedora)
* RPMfind.net (Various RPM-based distributions, indexed by category)
Updates:
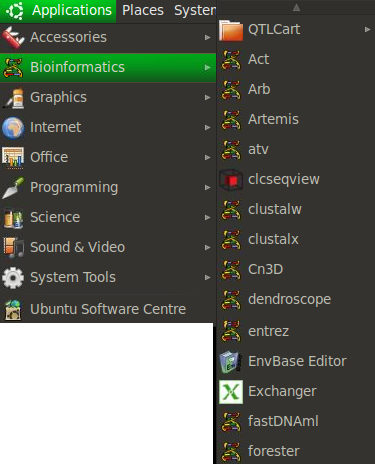
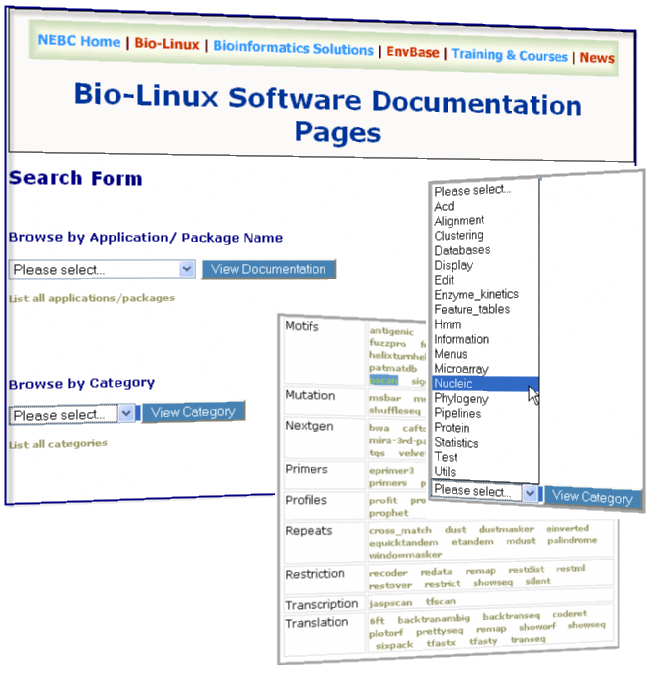
Bio-Linux 6.0 is a 64-bit workstation system developed for biologists and bioinformaticians by the NERC Environmental Bioinformatics Centre. Bio-Linux 6.0 is based on the popular Ubuntu Linux system, giving you the benefits of a free, user-friendly, comprehensive computing environment, as well as the convenience of the bioinformatics tools you need. Bio-Linux 6.0 comes with over 500 bioinformatics programs pre-installed, including software for handling sequence data from Roche 454 and Illumina systems, and R libraries of interest to biologists, such as vegan, ShortReads, Rsamtools, and the the biocLite selection of packages from Bioconductor. It also includes a comprehensive documentation system providing information on all the bioinformatics software on the system, categorised according to functionality. To help you get started, a folder of sample data is linked to the desktop, so you can try out new programs easily.
Installation of the system is simple, or you can boot directly from USB stick or DVD to run the system without installing anything to your hard disk. Bio-Linux 6.0 also comes with the FreenNX server, making it easier than ever to log in remotely from other machines and run a full, graphical Bio-Linux session. Of course, Bio-Linux also includes other customisations to make getting started on the system easy, while ensuring you are working in a secure environment.
Bio-Linux 6.0 can be downloaded from the NEBC website, and used to create a bootable USB stick or DVD. You can then either run the system live from either of these, or install the system. Full details, including links to download the new system, can be found on our website: http://nebc.nerc.ac.uk/tools/bio-linux
Debian.
Debian is another very popular Linux distribution in use in many academic institutions, and some bioinformaticians have made their own software packages available for this distribution in the deb format.
* Debian Med (Debian contains a lot of medical software internally)
* NEBC Bio-Linux (Non-standard Debian)
Apple/Mac
Many Linux packages are compatible with Mac OS X and there are several projects which attempt to make it easy to install selected Linux packages (including bioinformatics software) on a computer running Mac OS X. These include:
* Fink scientific packages
* Homebrew
Similarly, eBioinformatics provides a Mac OS GUI for over 300 open source bioinformatics programs.
You can install Bio-Linux on your machine, either as the only operating system, or as part of a dual-boot setup which allows you to use your current system and Bio-Linux on the same hardware.
Bio-Linux also runs Live from the DVD. This runs in the memory of your machine and does not involve installing anything. This is a great, no-hassle way to try out Bio-Linux, demonstrate or teach with it, or to work with when you are on the move.
Getting Bio-Linux.
Getting Bio-Linux is simple. Just download the image, burn it to a DVD, and boot the machine from that DVD. See this page for troubleshooting help regarding the DVD.
Bio-Linux can also be run from a USB memory stick. This is an ideal way to work with Bio-Linux Live, as files you create are saved to the stick. Of course, you can install the system from the memory stick also.
You can easily create bootable memory sticks yourself using the bio-linux-usb-maker package.
Until recently, you could request a Bio-Linux on a 4.0Gb memory stick if you were funded by the NERC or have a working relationship with the NEBC. This service will be offered again, after the release of Bio-Linux 6.0 in June 2010.
You can install our software packages onto a pre-existing Debian or Ubuntu system.
You can set up the system in various configurations depending on your needs.
You can submit feedback or ideas for Bio-Linux using our feedback form.
High throughput sequence data handling on Bio-Linux
Screenshots.









0 commenti:
Post a Comment